Regulation of replication origin activation
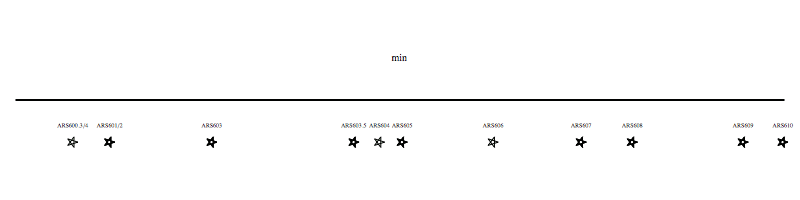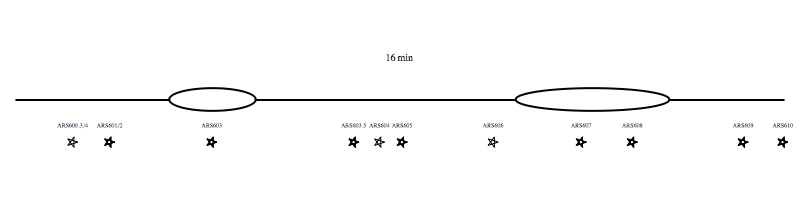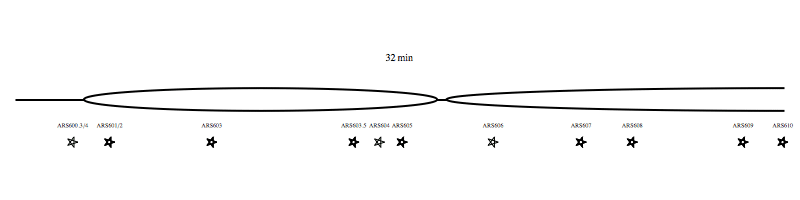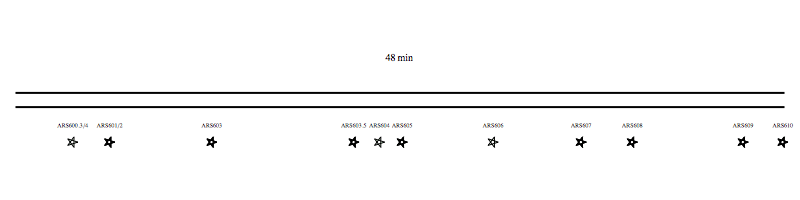
Each region of the genome replicates at a characteristic time, determined by the time that the replication origins activate. We have developed genomic approaches to measure origin activation and the times at which regions of the genome replicate. However, within individual cells replication origins activate stocastically, therefore each cell use a different combination of origins to replication the genome. The video clip (below) shows an example of one possible scenario for the replication of chromosome VI. We are interested in understanding what regulates the activity of origins and how the cell ensures that sufficient, appropriately distributed, origins are activated to complete genome replication in the time available.
Each region of the genome replicates at a characteristic time, determined by the time that the replication origins activate. We have developed genomic approaches to measure origin activation and the times at which regions of the genome replicate. However, within individual cells replication origins activate stocastically, therefore each cell use a different combination of origins to replication the genome. The video clip (above) shows an example of one possible scenario for the replication of chromosome VI. We are interested in understanding what regulates the activity of origins and how the cell ensures that sufficient, appropriately distributed, origins are activated to complete genome replication in the time available.
Related publications
High-resolution replication profiles define the stochastic nature of genome replication initiation and termination.
Hawkins et al. (2013)
Cell Rep, 5(4):1132-41
Conservation of replication timing reveals global and local regulation of replication origin activity.
Müller & Nieduszynski (2012)
Genome Res., 22(10):1953-62
Dynamics of DNA replication in yeast.
Retkute et al. (2011)
Phys. Rev. Lett., 107(6):068103
Mathematical modelling of whole chromosome replication.
de Moura et al. (2010)
Nucleic Acids Res., 38(17):5623-33